PROJECT DETAILS
- Category Early Research
The goal of this research is to develop population genetic models to provide an understanding of mechanisms by which abiotic and biotic interactions generate selection on dispersal.
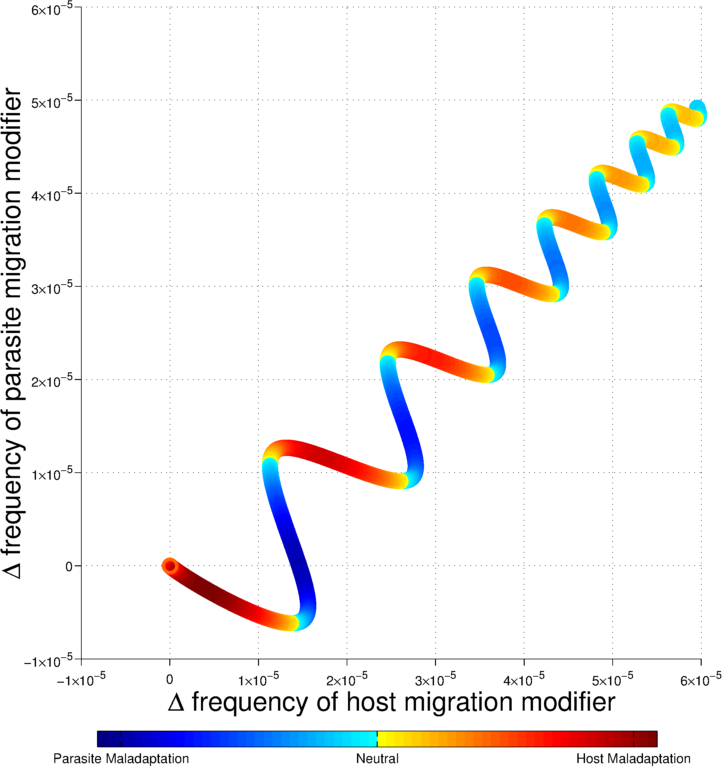
In populations at an adaptive equilibrium with their local environment, selection should favor philopatry over dispersal to different environments. But, theory shows that spatial environmental variation alone selects against dispersal. Yet, the vast majority of species exhibit some level of dispersal, mediated by behavior or morphology. Parasites are a particularly puzzling example. Many parasites are highly mobile, use different hosts within a life cycle, and have broad host ranges.
In natural populations in many cases, instead of favoring philopatry, environmental variability appears to do the opposite: it favors higher dispersal rates as a “bet hedging” strategy. Increased dispersal rates can be favored when particular forms of spatial variability are combined with temporal variability in fitness. I have found that coevolution of antagonistic interactions generates copious spatial variability in fitness in much the same way that host-parasite arms races generate temporal variability in fitness in models of the evolution of sex and recombination. In Drown, Dybdahl and Gomulkiewicz (2013), I show that antagonistic host-pathogen interactions generate both spatial and temporal fluctuations in fitness and thus favor increased dispersal rates. Under certain conditions, where interactions are strong, I find that there is a continuous evolutionary escalation in dispersal rates, which contributes to explaining the ubiquity of dispersal in nature . The long term evolution of dispersal described by my model provides an understanding of the empirical evidence of local adaptation necessary for host-parasite interactions to drive the evolution of host and pathogen dispersal.
- Drown, D. M., Dybdahl, M. F., & Gomulkiewicz, R. (2013). Consumer-resource interactions and the evolution of migration. Evolution, 67(11), 3290-3304. https://doi.org/10.1111/evo.12194
The goal of this research is to develop evolutionary genetic theory for understanding how coevolution between hosts and their symbionts affects co-transmission of host and symbiont genomes.
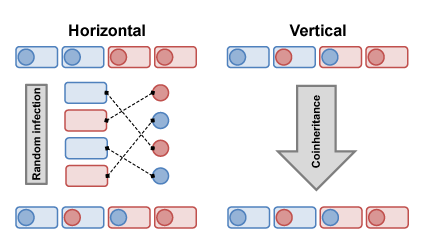
Intimate host-symbiont associations are ubiquitous in natural communities. Moreover, the evolutionary merger of genomes from different species or kingdoms has facilitated major innovations in eukaryotic life. However, the dual control of a single phenotype by two genomes generates conflicts and inefficiencies. Persistent or obligate associations require mechanisms that insure that symbionts are transmitted from one host to another each generation and that insure coordinated trans-genomic gene expression. The mode of symbiont transmission ranges from vertical transmission (parents to offspring) at one extreme to horizontal (contagious) transmission at the other. The mode of transmission appears to be phylogenetically labile, much like plant mating systems. I am trying to understand the evolutionary forces which cause transitions from one mode to the other.
I am developing population genetic models and computer simulations to investigate the evolution of symbiont transmission. While it might seem intuitive that vertical transmission would be selected for by an obligate host-symbiont interaction, I have found that, without stochastic forces like mutation, conditions favoring vertical transmission of an obligate symbiont are restrictive (Drown et al. 2013). In the absence of epistatic fitness effects between host and symbiont genes, the genetic covariance between host and symbiont created by vertical transmission is quickly broken down by even very limited amounts of horizontal transmission. My theoretical work shows that such intimate host-symbiont coevolution requires a high degree of specificity of the fitness interactions between both players.
As an example of the potential of my theory research, I have recently applied the theory of co-transmission of genomes to understanding the distribution of genes within genomes. Mitochondria, being vertically transmitted, serve as an excellent case study. In Drown, Preuss and Wade (2012), we discovered that nuclear genes which interact with the mitochondrion are significantly under-represented on the X-chromosome in mammals. This study links evolutionary theory of sexual conflict and genomic conflict to explain a highly significant and broad scale genomic pattern.
- Drown, D. M., Preuss, K. M., & Wade, M. J. (2012). Evidence of a paucity of genes that interact with the mitochondrion on the X in mammals. Genome Biology and Evolution, 4(8), 875-880. https://doi.org/10.1093/gbe/evs064